Usage
For best practice, output from GAPIT should be in its own folder. In this case, they are located in a folder called GWAS_Results/. For this example we will plot GWAS results from 3 traits in a lentil diversity panel:
- ****Cotyledon_Color**: a qualitative trait describing cotyledon color (Red = 0, Yellow = 1).
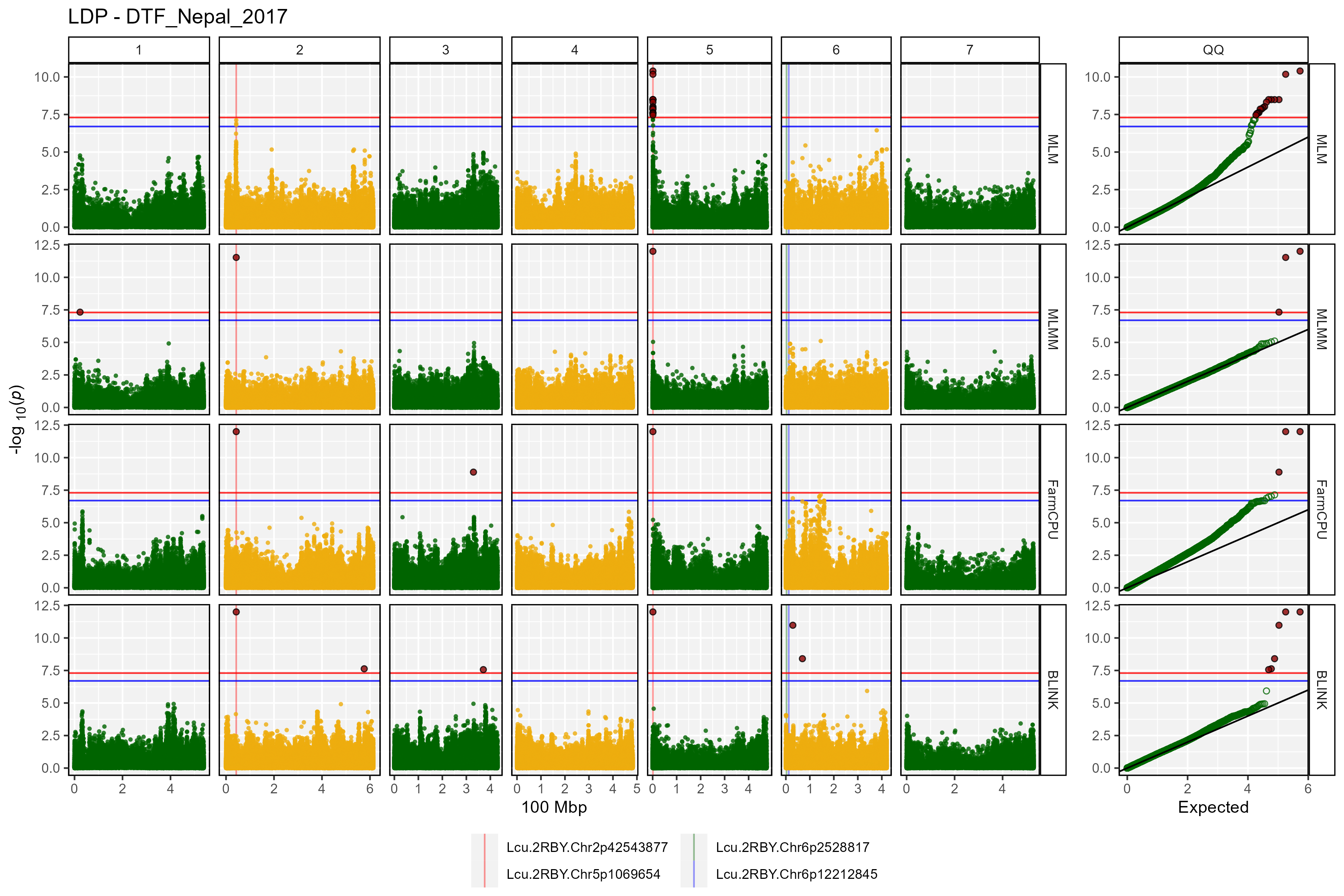
- DTF_Nepal_2017: a quantitative trait describing days from sowing to flowering in a 2017 Nepal field trial.
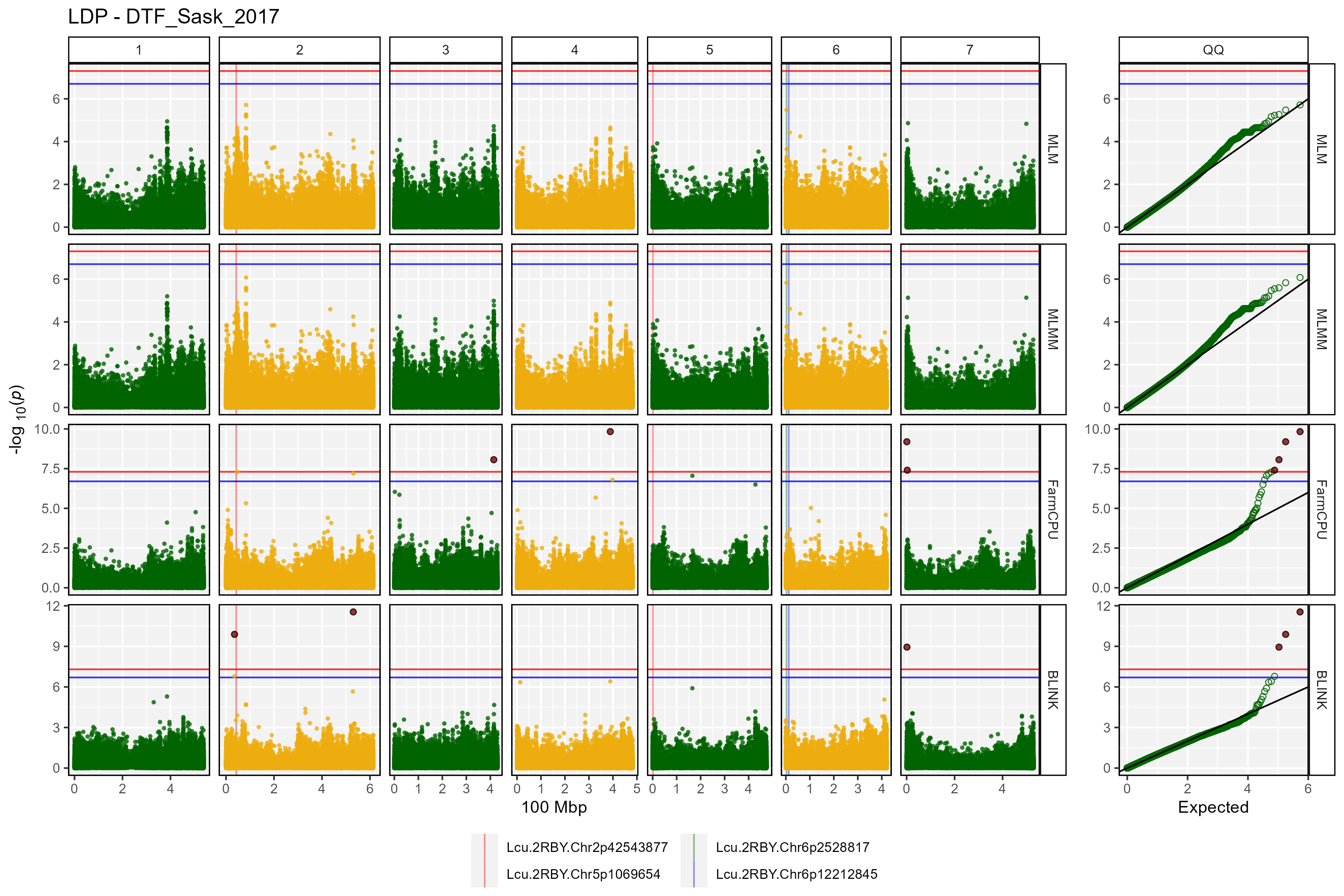
- DTF_Sask_2017: a quantitative trait describing days from sowing to flowering in a 2017 Saskatchewan field trial.
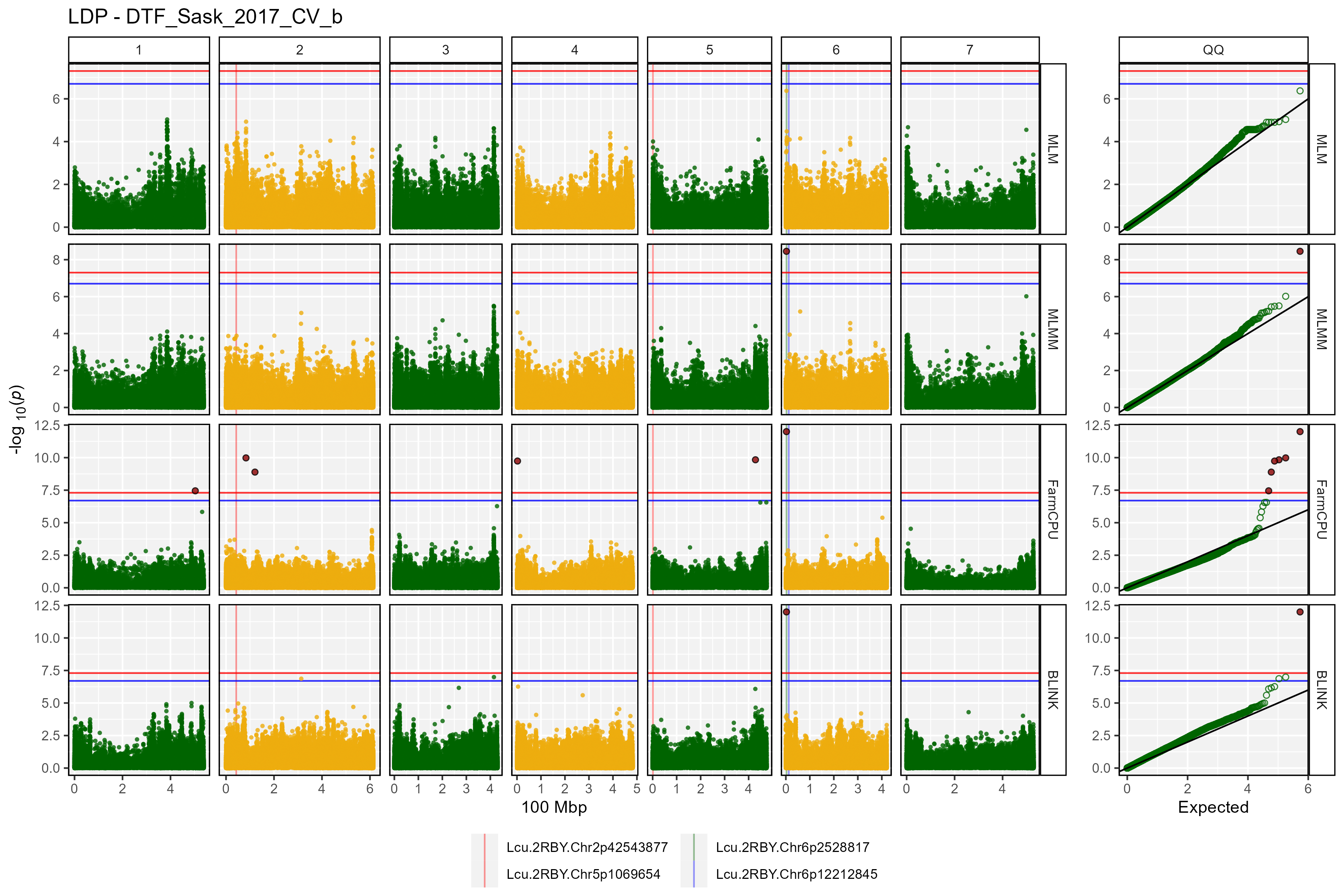
- DTF_Sask_2017_b: same as above but run with the b coefficient from a photothermal model (see Wright et al. 2020) used as a covariate.
Note: for more info check out this GWAS tutorial.
List Traits
myTraits <- list_Traits(folder = "GWAS_Results/")
myTraitsList Results Files
myFiles <- list_Result_Files(folder = "GWAS_Results/")
myFiles## [1] "GAPIT.Association.GWAS_Results.BLINK.Cotyledon_Color(Kansas).csv"
## [2] "GAPIT.Association.GWAS_Results.BLINK.Cotyledon_Color(NYC).csv"
## [3] "GAPIT.Association.GWAS_Results.BLINK.DTF_Nepal_2017(Kansas).csv"
## [4] "GAPIT.Association.GWAS_Results.BLINK.DTF_Nepal_2017(NYC).csv"
## [5] "GAPIT.Association.GWAS_Results.BLINK.DTF_Sask_2017(Kansas).csv"
## [6] "GAPIT.Association.GWAS_Results.BLINK.DTF_Sask_2017(NYC).csv"
## [7] "GAPIT.Association.GWAS_Results.BLINK.DTF_Sask_2017_b(Kansas).csv"
## [8] "GAPIT.Association.GWAS_Results.BLINK.DTF_Sask_2017_b(NYC).csv"
## [9] "GAPIT.Association.GWAS_Results.FarmCPU.Cotyledon_Color(Kansas).csv"
## [10] "GAPIT.Association.GWAS_Results.FarmCPU.Cotyledon_Color(NYC).csv"
## [11] "GAPIT.Association.GWAS_Results.FarmCPU.DTF_Nepal_2017(Kansas).csv"
## [12] "GAPIT.Association.GWAS_Results.FarmCPU.DTF_Nepal_2017(NYC).csv"
## [13] "GAPIT.Association.GWAS_Results.FarmCPU.DTF_Sask_2017(Kansas).csv"
## [14] "GAPIT.Association.GWAS_Results.FarmCPU.DTF_Sask_2017(NYC).csv"
## [15] "GAPIT.Association.GWAS_Results.FarmCPU.DTF_Sask_2017_b(Kansas).csv"
## [16] "GAPIT.Association.GWAS_Results.FarmCPU.DTF_Sask_2017_b(NYC).csv"
## [17] "GAPIT.Association.GWAS_Results.GLM.Cotyledon_Color(NYC).csv"
## [18] "GAPIT.Association.GWAS_Results.GLM.DTF_Nepal_2017(NYC).csv"
## [19] "GAPIT.Association.GWAS_Results.GLM.DTF_Sask_2017(NYC).csv"
## [20] "GAPIT.Association.GWAS_Results.GLM.DTF_Sask_2017_b(NYC).csv"
## [21] "GAPIT.Association.GWAS_Results.MLM.Cotyledon_Color(NYC).csv"
## [22] "GAPIT.Association.GWAS_Results.MLM.DTF_Nepal_2017(NYC).csv"
## [23] "GAPIT.Association.GWAS_Results.MLM.DTF_Sask_2017(NYC).csv"
## [24] "GAPIT.Association.GWAS_Results.MLM.DTF_Sask_2017_b(NYC).csv"
## [25] "GAPIT.Association.GWAS_Results.MLMM.Cotyledon_Color(NYC).csv"
## [26] "GAPIT.Association.GWAS_Results.MLMM.DTF_Nepal_2017(NYC).csv"
## [27] "GAPIT.Association.GWAS_Results.MLMM.DTF_Sask_2017(NYC).csv"
## [28] "GAPIT.Association.GWAS_Results.MLMM.DTF_Sask_2017_b(NYC).csv"Check Results
is_ran(folder = "GWAS_Results/")List Significant Markers
# first reorder the result files if they are not already arranged by P.value
order_GWAS_Results(folder = "GWAS_Results/", files = myFiles)
is_ordered(folder = "GWAS_Results/")## Trait GLM MLM MLMM FarmCPU BLINK CMLM SUPER FarmCPU_Kansas
## 1 Cotyledon_Color X X X X X NA NA X
## 2 DTF_Nepal_2017 X X X X X NA NA X
## 3 DTF_Sask_2017 X X X X X NA NA X
## 4 DTF_Sask_2017_b X X X X X NA NA X
## BLINK_Kansas
## 1 X
## 2 X
## 3 X
## 4 X
myResults <- table_GWAS_Results(folder = "GWAS_Results/", fnames = myFiles,
threshold = 6.8, sug.threshold = 5)
myResults[1:10,]## SNP Chr Pos P.value MAF effect
## 1 Lcu.1GRN.Chr1p365986872 1 365986872 1.754402e-175 0.47096774 0.4683785
## 2 Lcu.1GRN.Chr1p365986872 1 365986872 1.164052e-154 0.47096774 NA
## 3 Lcu.1GRN.Chr1p365986872 1 365986872 3.281941e-128 0.47096774 0.4954384
## 4 Lcu.1GRN.Chr1p361840399 1 361840399 2.178943e-84 0.04193548 0.4603589
## 5 Lcu.1GRN.Chr1p361840399 1 361840399 6.963600e-47 0.04193548 0.4715544
## 6 Lcu.1GRN.Chr1p361856257 1 361856257 3.377658e-40 0.04516129 NA
## 7 Lcu.1GRN.Chr1p365986872 1 365986872 2.845683e-37 0.47096774 NA
## 8 Lcu.1GRN.Chr1p365318023 1 365318023 1.769760e-35 0.47096774 NA
## 9 Lcu.1GRN.Chr1p365318027 1 365318027 2.803987e-35 0.47258065 NA
## 10 Lcu.1GRN.Chr5p1658484 5 1658484 1.050791e-34 0.12345679 12.5619150
## H.B.P.Value Model Type Trait negLog10_P negLog10_HBP
## 1 5.901229e-170 BLINK Kansas Cotyledon_Color 174.75587 169.22906
## 2 3.915488e-149 MLMM NYC Cotyledon_Color 153.93403 148.40721
## 3 1.103937e-122 FarmCPU Kansas Cotyledon_Color 127.48387 121.95706
## 4 3.664623e-79 BLINK Kansas Cotyledon_Color 83.66175 78.43597
## 5 1.171163e-41 FarmCPU Kansas Cotyledon_Color 46.15717 40.93138
## 6 5.680663e-35 MLMM NYC Cotyledon_Color 39.47138 34.24560
## 7 9.571939e-32 GLM NYC Cotyledon_Color 36.54581 31.01900
## 8 2.976445e-30 GLM NYC Cotyledon_Color 34.75209 29.52630
## 9 3.143895e-30 GLM NYC Cotyledon_Color 34.55222 29.50253
## 10 3.534516e-29 BLINK Kansas DTF_Nepal_2017 33.97848 28.45167
## Threshold Effect
## 1 Significant NA
## 2 Significant 0.4905608
## 3 Significant NA
## 4 Significant NA
## 5 Significant NA
## 6 Significant 1.4947330
## 7 Significant 0.4558343
## 8 Significant 0.4470957
## 9 Significant 0.4451259
## 10 Significant NA
list_Top_Markers(folder = "GWAS_Results/", trait = "DTF_Nepal_2017",
threshold = 6.7, chroms = c(2,5))## # A tibble: 8 × 6
## SNP Chr Pos Traits Models Max_LogP
## <chr> <int> <int> <chr> <chr> <dbl>
## 1 Lcu.1GRN.Chr5p1658484 5 1658484 DTF_Nepal_2017 BLINK; FarmCPU;… 34.0
## 2 Lcu.1GRN.Chr2p44546658 2 44546658 DTF_Nepal_2017 FarmCPU; GLM; M… 33.0
## 3 Lcu.1GRN.Chr2p44545877 2 44545877 DTF_Nepal_2017 BLINK; FarmCPU;… 22.7
## 4 Lcu.1GRN.Chr2p44558948 2 44558948 DTF_Nepal_2017 GLM; MLM 22.2
## 5 Lcu.1GRN.Chr5p1650591 5 1650591 DTF_Nepal_2017 FarmCPU; GLM; M… 18.7
## 6 Lcu.1GRN.Chr5p2101990 5 2101990 DTF_Nepal_2017 FarmCPU 14.8
## 7 Lcu.1GRN.Chr5p1651791 5 1651791 DTF_Nepal_2017 GLM; MLM 14.1
## 8 Lcu.1GRN.Chr5p2102310 5 2102310 DTF_Nepal_2017 FarmCPU 7.40
list_Top_Markers(folder = "GWAS_Results/", trait = "DTF_Sask_2017_b",
threshold = 6.7, chroms = 6)## # A tibble: 9 × 6
## SNP Chr Pos Traits Models Max_LogP
## <chr> <int> <int> <chr> <chr> <dbl>
## 1 Lcu.1GRN.Chr6p3269280 6 3269280 DTF_Sask_2017_b BLINK; FarmC… 32.9
## 2 Lcu.1GRN.Chr6p1734191 6 1734191 DTF_Sask_2017_b GLM 23.6
## 3 Lcu.1GRN.Chr6p1445607 6 1445607 DTF_Sask_2017_b GLM 23.5
## 4 Lcu.1GRN.Chr6p3270522 6 3270522 DTF_Sask_2017_b FarmCPU 21.8
## 5 Lcu.1GRN.Chr6p431657465 6 431657465 DTF_Sask_2017_b FarmCPU 17.8
## 6 Lcu.1GRN.Chr6p324049462 6 324049462 DTF_Sask_2017_b BLINK 13.1
## 7 Lcu.1GRN.Chr6p46982948 6 46982948 DTF_Sask_2017_b BLINK 9.10
## 8 Lcu.1GRN.Chr6p431595092 6 431595092 DTF_Sask_2017_b FarmCPU 9.04
## 9 Lcu.1GRN.Chr6p172232372 6 172232372 DTF_Sask_2017_b FarmCPU 8.71
list_Top_Markers(folder = "GWAS_Results/", trait = "Cotyledon_Color",
threshold = 6.7, chroms = 6, n = 2)## # A tibble: 2 × 6
## SNP Chr Pos Traits Models Max_LogP
## <chr> <int> <int> <chr> <chr> <dbl>
## 1 Lcu.1GRN.Chr6p297914010 6 297914010 Cotyledon_Color GLM 13.4
## 2 Lcu.1GRN.Chr6p303174618 6 303174618 Cotyledon_Color GLM 12.8
myMarkers <- c("Lcu.1GRN.Chr2p44545877", "Lcu.1GRN.Chr5p1658484",
"Lcu.1GRN.Chr6p3269280", "Lcu.1GRN.Chr1p365986872")Manhattan Plots
Multi Manhattan Plots
for(i in myTraits) {
mp <- gg_Manhattan(folder = "GWAS_Results/",
trait = i,
title = paste("LDP -", i),
threshold = 6.7,
sug.threshold = 5,
vlines = myMarkers,
vline.colors = c("red","red","darkgreen","blue"),
vline.types = c(1,1,1,1),
vline.legend = T,
facet = F,
addQQ = T,
pmax = 12,
models = c("MLM", "MLMM", "FarmCPU", "BLINK"),
model.colors = c("darkgreen", "darkred", "darkorange3", "steelblue"),
legend.rows = 2)
ggsave(paste0("man/figures/Multi_", i, ".png"),
mp, width = 12, height = 4, bg = "white")
}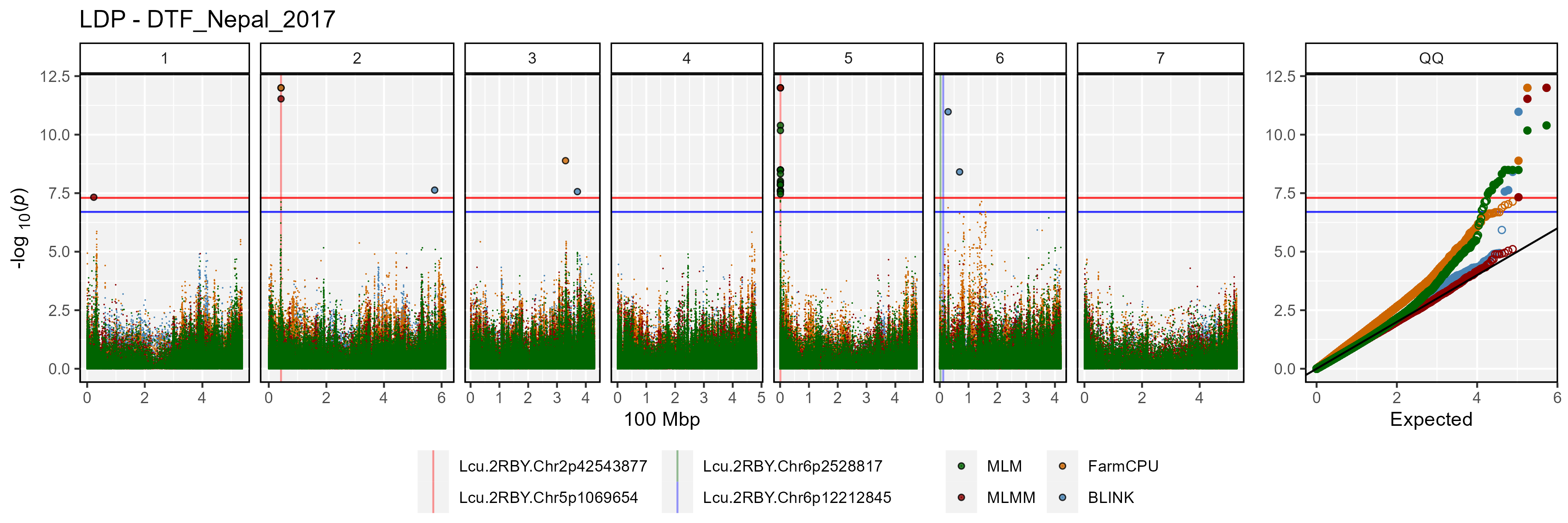
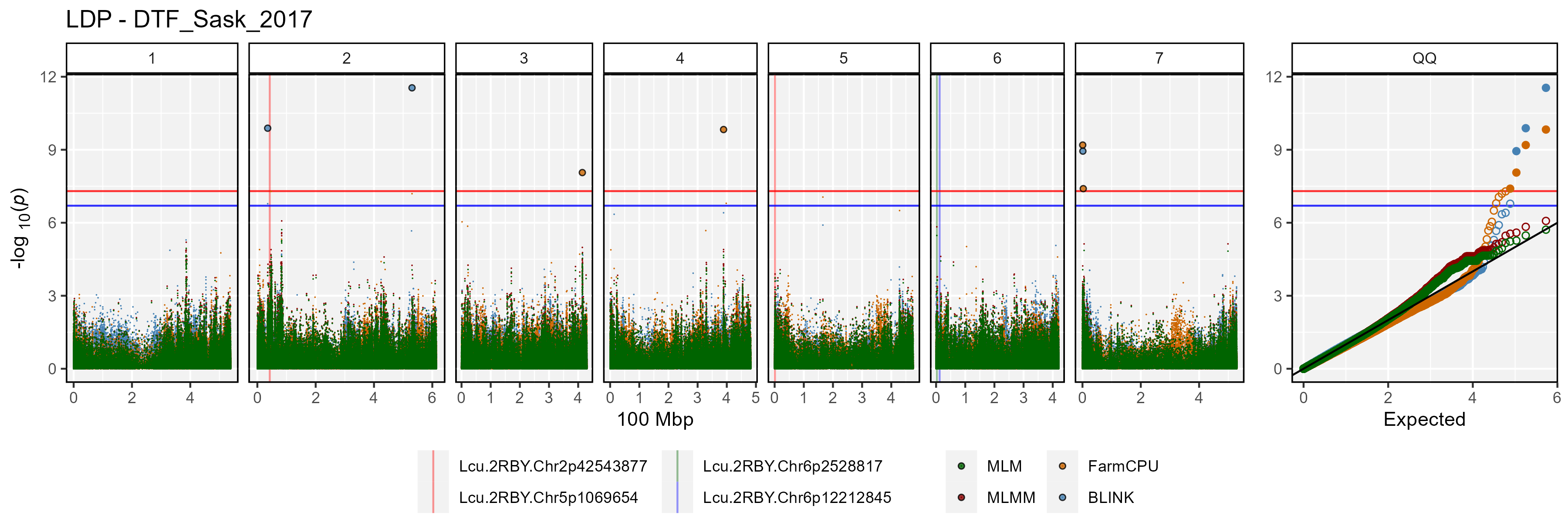
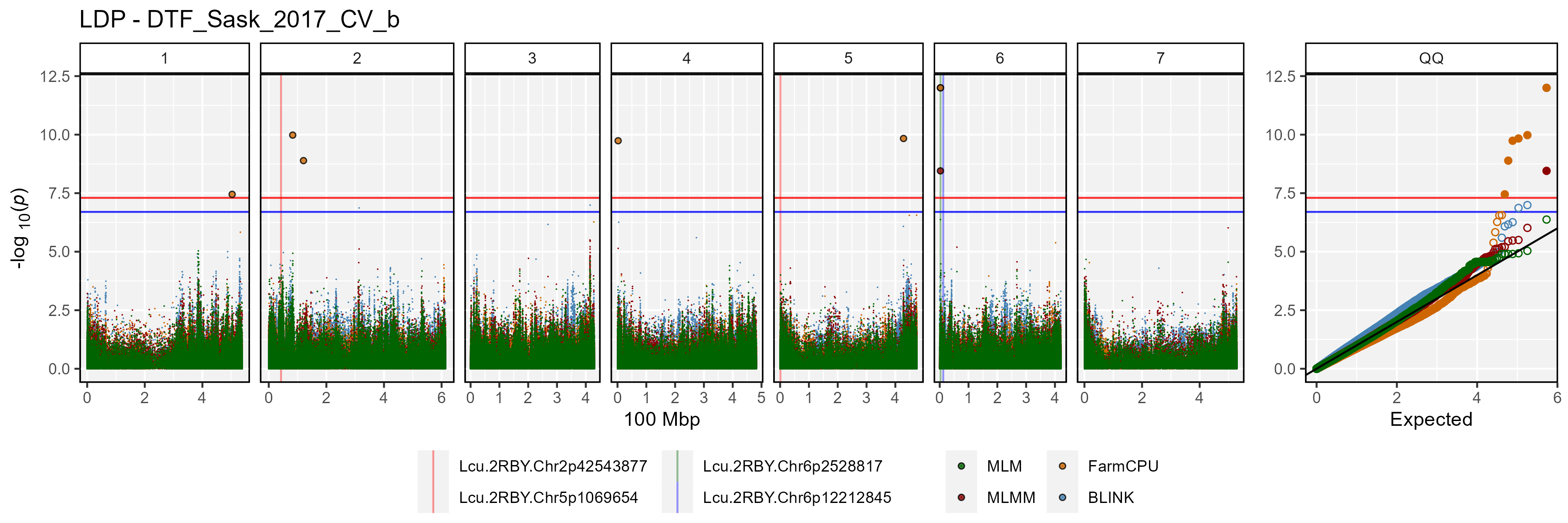
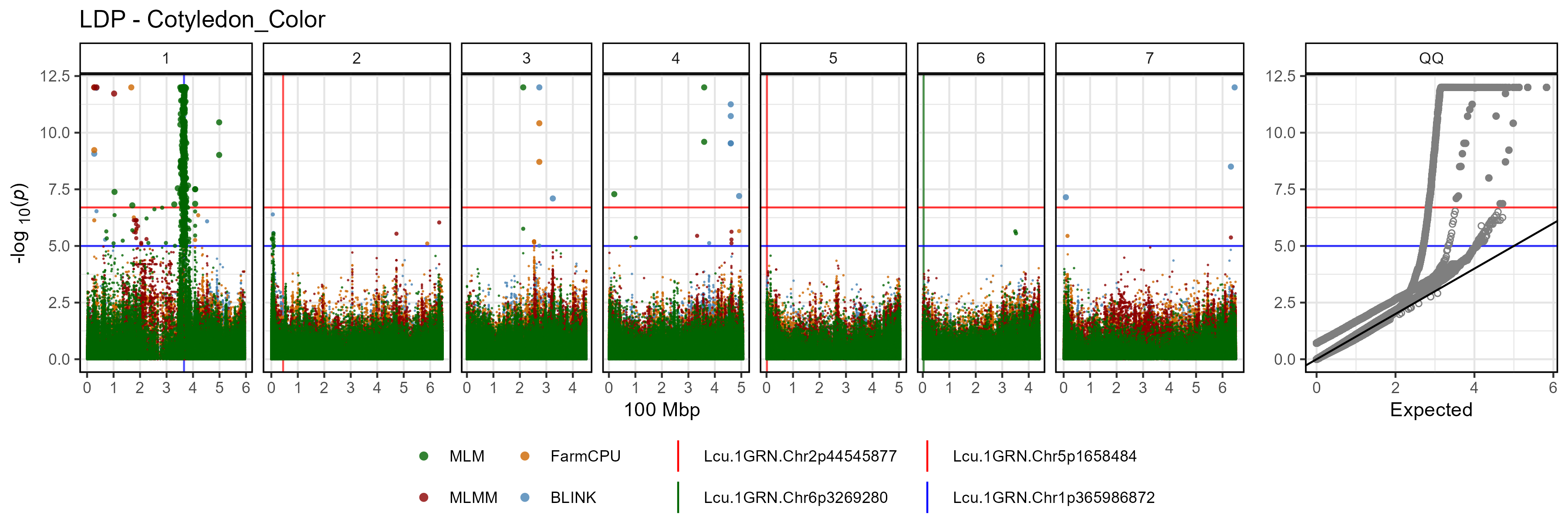
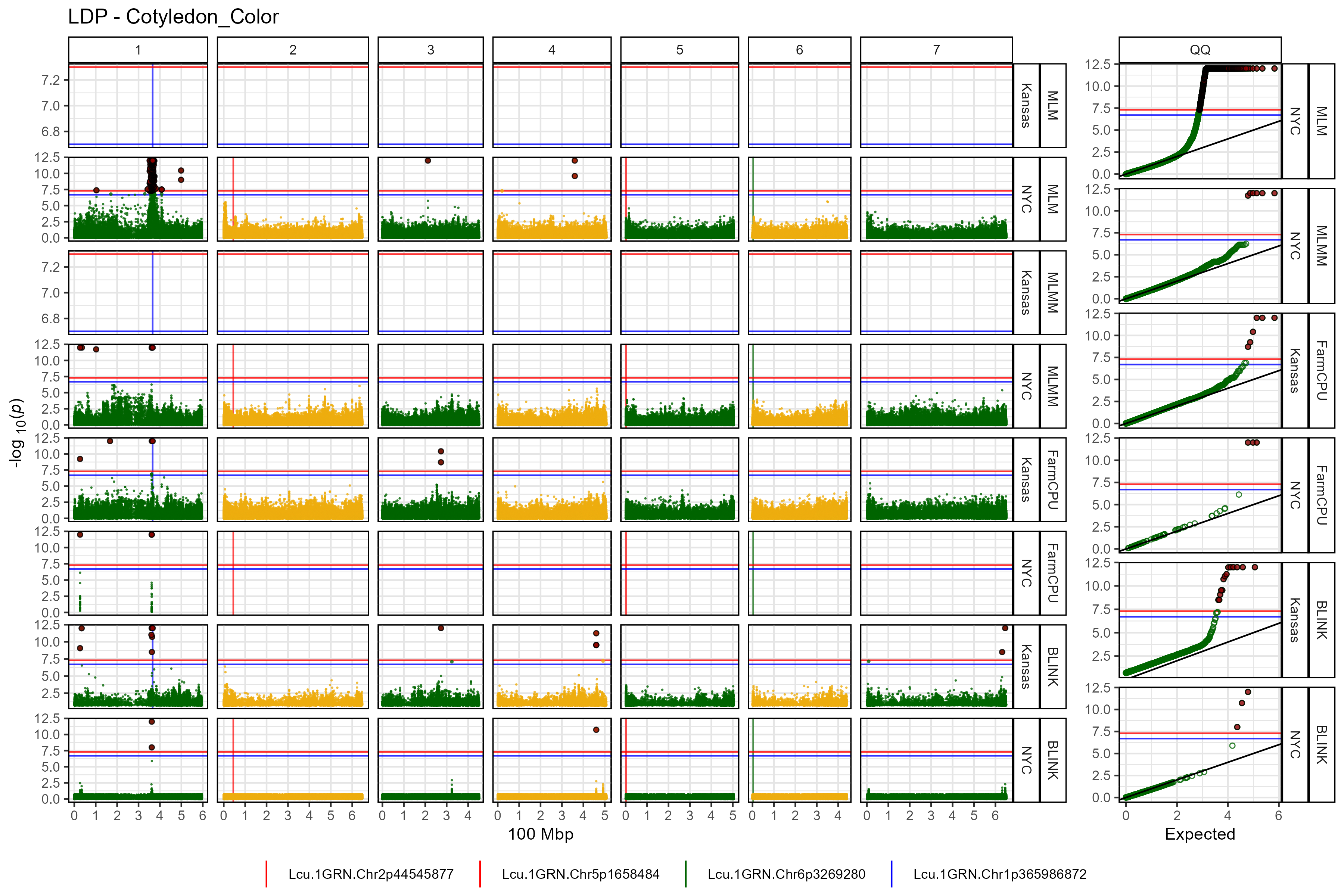
Facetted Manhattan Plots
for(i in myTraits) {
mp <- gg_Manhattan(folder = "GWAS_Results/",
trait = i,
title = paste("LDP -", i),
threshold = 7.3,
sug.threshold = 6.7,
vlines = myMarkers,
vline.colors = c("red","red","darkgreen","blue"),
vline.types = c(1,1,1,1),
vline.legend = T,
facet = T,
addQQ = T,
pmax = 12,
models = c("MLM", "MLMM", "FarmCPU", "BLINK"),
chrom.colors = rep(c("darkgreen", "darkgoldenrod2"), 4),
legend.rows = 1)
ggsave(paste0("man/figures/Facet_", i, ".png"),
mp, width = 12, height = 8, bg = "white")
}
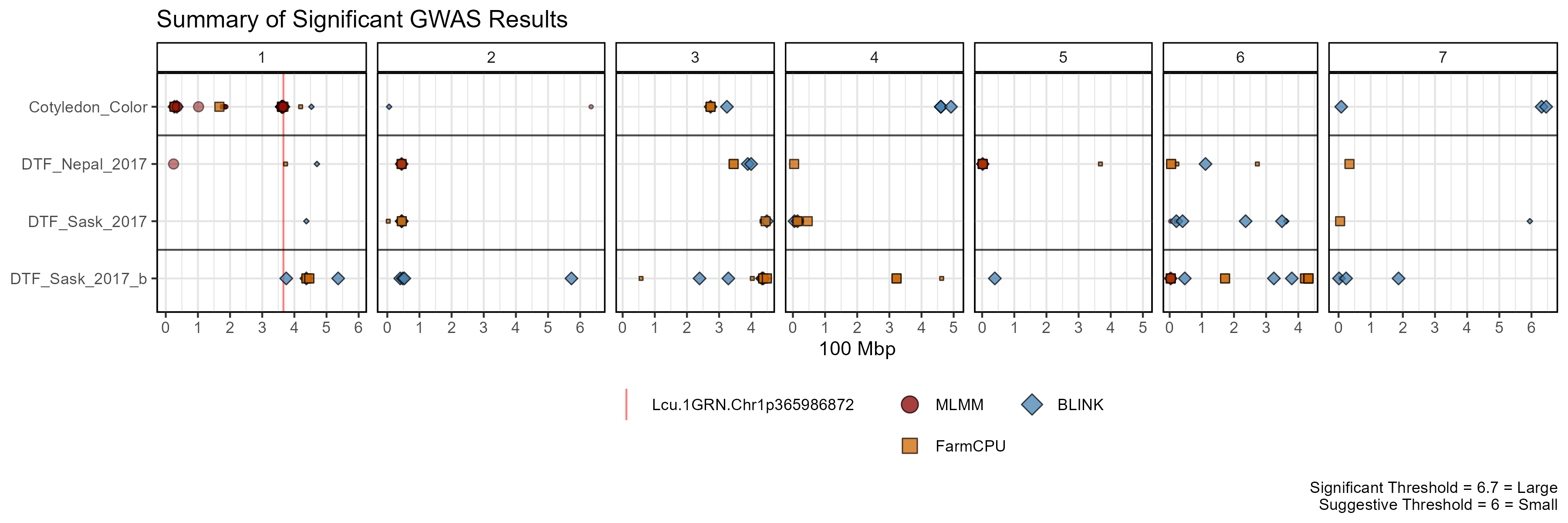
Summary Plot
mp <- gg_GWAS_Summary(folder = "GWAS_Results/",
traits = myTraits,
models = c("MLM", "MLMM", "FarmCPU", "BLINK"),
colors = c("darkgreen", "darkred", "darkorange3", "steelblue"),
threshold = 6.7, sug.threshold = 6,
hlines = c(1.5,3.5), legend.rows = 2,
vlines = myMarkers,
vline.colors = c("red", "red", "green", "blue"),
vline.types = c(1,1,1,1),
title = "Summary of Significant GWAS Results")
ggsave("man/figures/GWAS_Summary_01.png", mp, width = 12, height = 4)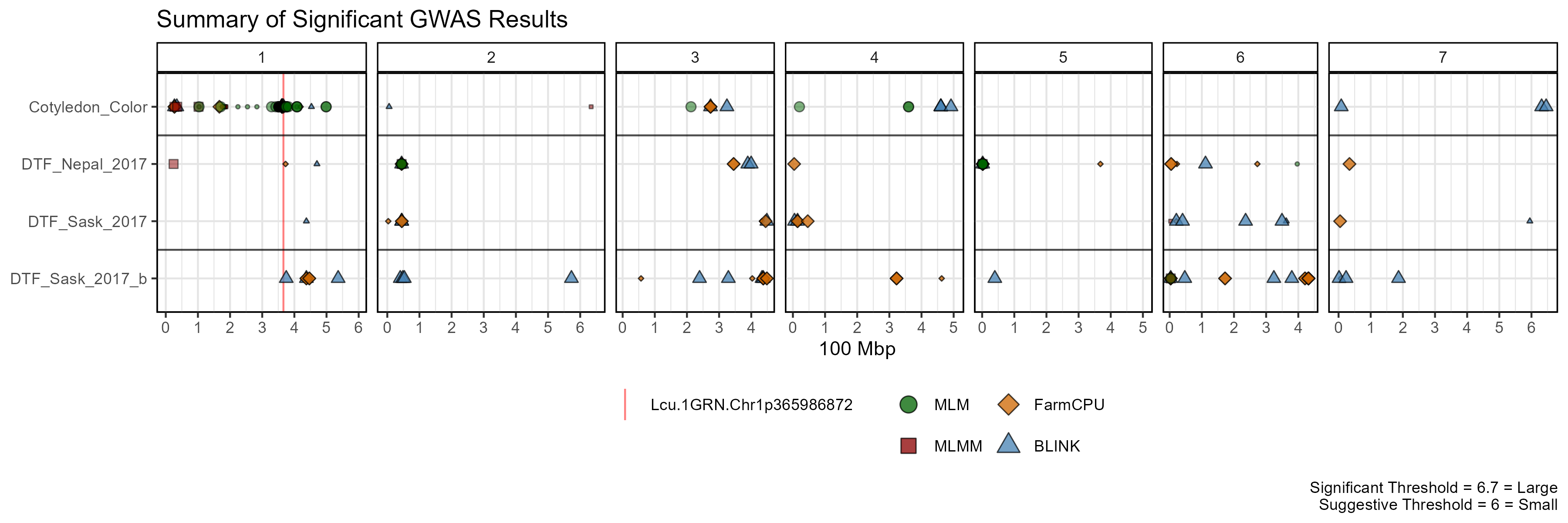
mp <- gg_GWAS_Summary(folder = "GWAS_Results/",
traits = myTraits,
models = c("MLMM", "FarmCPU", "BLINK"),
colors = c("darkred", "darkorange3", "steelblue"),
threshold = 6.7, sug.threshold = 6,
hlines = c(1.5,3.5), legend.rows = 2,
vlines = myMarkers,
vline.colors = c("red", "red", "green", "blue"),
vline.types = c(1,1,1,1),
title = "Summary of Significant GWAS Results")
ggsave("man/figures/GWAS_Summary_02.png", mp, width = 12, height = 4)